Segmenting cells
To extract individual cells from the probability image, use CellFinder class from cloudmap.ecc.cellfinder_parallel.
Example script
from ecc.cellfinder import CellFinder
# create a new instance
cf = CellFinder()
# force it print detailed progress
cf.set_verbose(True)
# Optional: set number of parallel CPU cores
cf.set_num_workers(4)
# Optional: set block size
cf.set_block_size({"blocksize": 120, "overlap": 20})
# set voxel size of the input image
cf.set_image_voxel_size( {"X": 6.45, "Y": 6.45, "Z": 7.0} )
# set some parameters
cf.set_prob_threshold(0.70)
cf.set_min_particle_volume(2)
cf.set_max_particle_volume(64)
cf.set_intensity_computation_mode('obj_mean')
# Optional: give it a nice nickname
cf.set_nickname('test')
# set output directory
cf.set_outdir('/data/cellfinder-result/')
cf.set_raw_image_path('/data/test-image.hdf5')
cf.set_prob_image_path('/data/classifier-result/prob_image.h5')
# run!
cf.run_main()
Tips:
To process multiple brains, add for loop in the above code.
Some more explanation…
Create a new instance
from ecc.cellfinder import CellFinder
cf = CellFinder()
(Optional) Set the number of parallel threads
You can control the number of threads allocated for this computation by
cf.set_num_workers(4)
By default, ecc will try to use all of the CPU cores available on the system.
(Optional) Set verbose
By telling set_verbose(True), the detailed progress information is printed on the screen output. If you want a silent execution, give it a false value.
cf.set_verbose(True)
(Optional) Set blocksizes and overlaps
CellFinder divides the large 3D into small blocks and processes them in a parallel manner. You can adjust the block size and overlap value by
cf.set_block_size({"blocksize": 120, "overlap": 20})
The default is {"blocksize": 120, "overlap": 20}, and you normally do not need to change this value.
Set voxel size of the input image
First, set voxel size of the input image sin μm:
cf.set_image_voxel_size( {"X": 6.45, "Y": 6.45, "Z": 7.0} )
If you want silent execution, give verbose false value.
CAUTION
Do not put wrong voxel size here, or all subsequent analysis will fail!!
Set counting paramters
cf.set_prob_threshold(0.70)
cf.set_min_particle_volume(2)
cf.set_max_particle_volume(64)
cf.set_intensity_computation_mode('obj_mean')
prob_threshold literally means the thresholding value applied to the probability image. Voxels with larger values than threshold are considred as objects (i.e. cell). The value should be in range [0, 1].
min_particle_volume defines the minimum volume (in voxel unit, not in μm3!) of the segmented objects. Objects smaller than this volume are ignored.
max_particle_volume defines the maximum volume (in voxel unit, not in μm3!) of the segmented objects. Objects larger than this volume is sent to object separation routine, where the objects gets separeted into multiple objects.
intensity_computation_mode defines how the intensity value is computed for each cell. Available options are max and obj_mean. max simply picks up the maximum intensity value of the segmented object. obj_mean computes the mean intensity value of the segmented object, calculated as
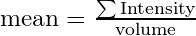
Namely, to recover the total intensity of the object, simply multiply as mean intensity with volume.
Set inputs and outpus
cf.set_nickname('test')
cf.set_outdir('/data/cellfinder-result/')
cf.set_raw_image_path('/data/test-image.hdf5')
cf.set_prob_image_path('/data/classifier-result/prob_image.h5')
nickname is used to name files.
All output files will be written in the directory you specified by cf.set_outdir().
Then, supply the paths to raw and probability image by calling cf.set_raw_image_path() and cf.set_prob_image_path().
(Optional) Set mask
Optionally, you can define a mask.
cf.set_mask_image_path( '/data/counting/mask.hdf5s' )
Image mask should be given as chunked HDF5 format. Voxels with values equal or greater than 1 are regarded as mask area, and voxels covered by the mask area are ignored.
For example, when strong autofluorescence is present on the brain surface (which is often the case), you can generate a brain surface mask to exclude those signals.
Run!
Now everything is ready. Start the calculation by
cf.run_main()
Tips:
Given 15GB image volume, this computation takes about 10 to 30 minutes using 30 CPU cores.
Inspect the output
In the output directory, you will find a csv table, named {nickname}_cells.csv, where nickname is the one that you defined.
In the table, each row will show the properties of one detected cell. The columns of this csv table will be
| X | Y | Z | deltaI | BG | vol | deltaI_total |
X, Y, Z are the center of the mass of the cell, following the coordinate convention defined here.
deltaI means the signal intensity, subtracted by background level. Depending on the intensity_computation_mode described above, it is either maximum or mean of the object.
BG is the background level. You can recover the original intensity by computing deltaI + BG.
vol is the volume of the cell.
deltaI_total is simply computed as deltaI * vol, corresponding to the total expression amount.